PLATFORM
Of the ~20,000 proteins in the human proteome, only about 600 have been successfully targeted with small molecule therapeutics. More than 5,000 undrugged proteins are known to be linked to disease, and more insights are needed on the remaining proteins to understand their function in human health. That leaves a vast proportion of the proteome unexplored and a big catalog of targets on drug developers’ wish lists.
See More
Know More
Drug Better
Belharra’s Searchlight™ Platform leverages our proprietary state-of-the art chemical probe library, specifically designed to map cryptic pockets, and a deep understanding of chemoproteomics to map the proteome and identify more informed starting points for groundbreaking medicines.
Searchlight Components
1
A computationally designed library of drug-like probes rapidly delivers non-covalent (reversible) starting points for new drugs
2
State-of-the-art photoaffinity-based chemoproteomics elucidates proteome- wide binding site locations
3
Cell-based screening identifies binding pockets in their native environment
4
Vast site-of-labeling datasets drive intelligent binding predictions and future library design
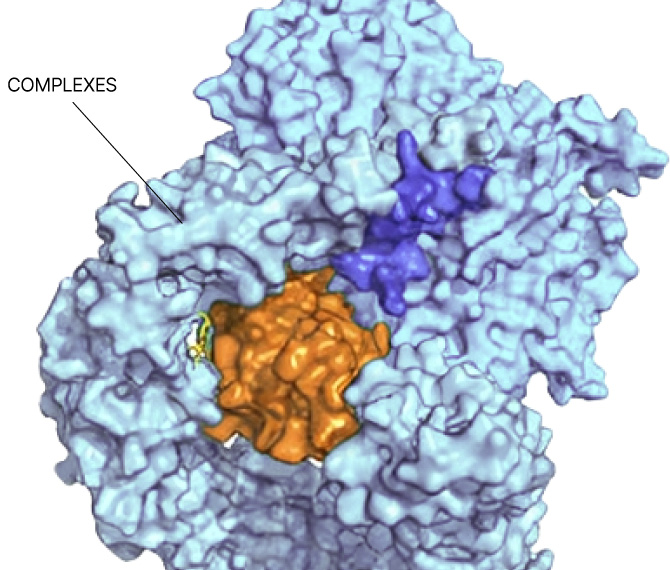
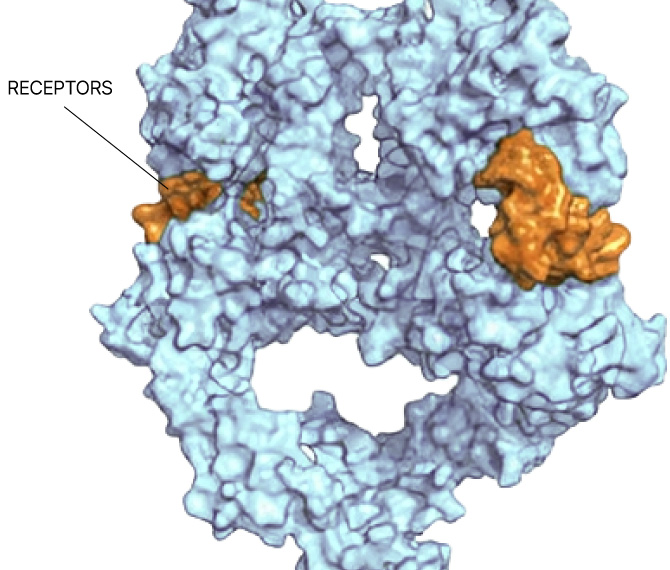
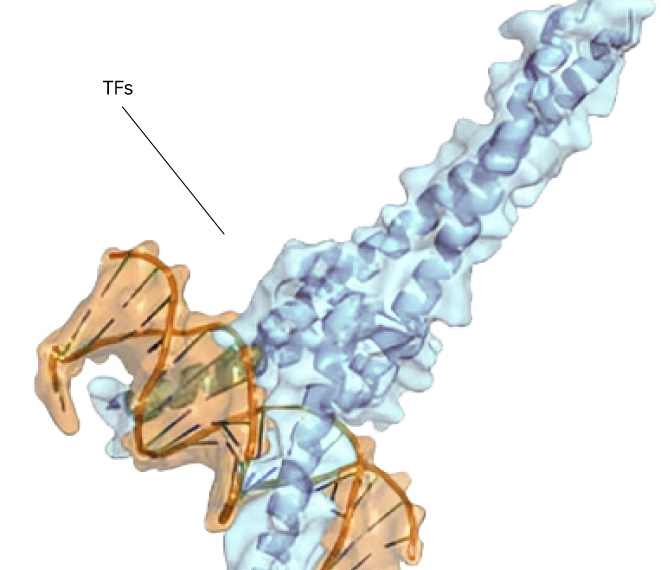
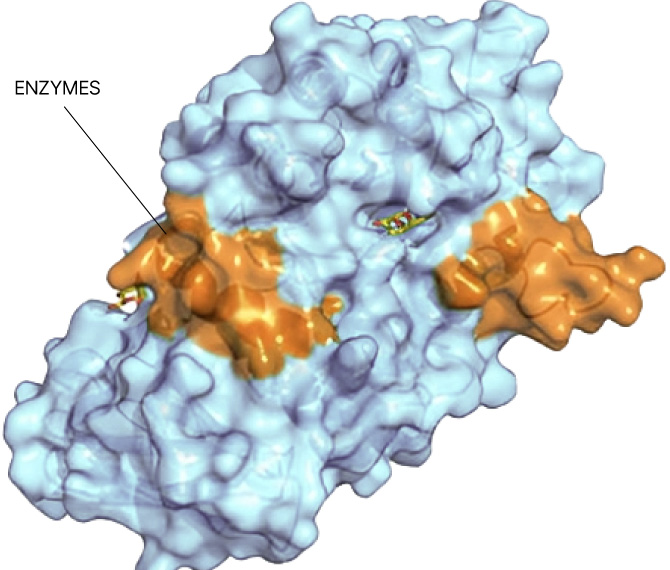
Small molecule drug discovery begins with finding a “binding pocket” on a protein. This pocket is where a small molecule can latch on and change the course of disease. Binding pockets can be active sites, allosteric sites, or at the interface of protein-protein interactions. But for the majority of proteins in the human proteome, no binding pocket has been found.
Our Searchlight Platform illuminates any binding pocket on any protein or protein complex, in any cell type, yielding insights that enable the discovery of new drugs for well-validated but elusive protein targets as well as novel drug targets.
Any site
On any protein
In any state
In any cell type
Searchlight leverages a proprietary, computationally designed library of non-fragment, drug-like molecules to “probe” the proteome and identify binding pockets. All screens are conducted in a native cellular environment to ensure that no binding pocket is missed. The platform leverages reversible (non-covalent) binding interactions, a differentiating feature versus other chemoproteomic platforms which rely on irreversible binding. The majority of approved medicines are non-covalent typically leading to a more tolerable safety profile and fewer risks for development.
See
more.
When a drug-like probe molecule reversibly binds to a protein, the Searchlight Platform uses photoaffinity chemistry to crosslink the probe to the bound protein. With exposure to ultraviolet light, the probe gets locked into place, enabling our team to identify which proteins are bound by the probe as well as the precise binding location on the target proteins. The site-of-labeling information allows us to rapidly understand the location and features of the pocket, greatly enabling medicinal chemistry optimization.
Know
more.
To date, our team has identified more than 4,000 binding pockets for the undrugged proteome. With these data and insights in hand, we have more informed, de-risked starting points to design and advance first-in-class and best-in-class medicines across a range of diseases and unmet patient needs. In addition, the proprietary proteome maps and site-of-labeling data are being leveraged to inform our machine learning and generative AI strategies and ultimately further augment platform performance.
Drug
better.
Next-Generation Chemoproteomics
Other chemoproteomics platforms
Designed to identify only covalent medicines with irreversible drug/protein binding
Enables identification of non-covalent medicines with reversible drug/protein binding (the majority of FDA-approved medicines are non-covalent)
Mainly fragment-based electrophilic probes that irreversibly bind cysteines
Non-fragment, computationally designed library of reversible, drug-like chemical probes
Typically relies on a ligandable cysteine being present
Photoaffinity labeling does not require a specific amino acid
Many screens conducted on isolated proteins
All screens conducted in native cellular environment, enabling the identification of pockets only present in live cells
Pocket identification is limited to specific sites, proteins, or cell types
Can find binding pockets on any site, on any protein, in any state, in any cell type
Our Focus
Our initial discovery pipeline includes well validated oncology and immunology targets. Complementing our internal discovery efforts, Belharra partners with industry-leading companies, lending the power of the Searchlight Platform to find new starting points for groundbreaking medicines targeting partner-identified proteins across all disease areas while growing the platform’s datasets.